The European Union aims to build a digital environment, the EU Digital Twin Ocean (EU DTO), that allows to create a digital replica of ocean processes to improve our understanding, predict their response to changes in the system, and to simulate alternative scenarios, which will ultimately lead to making better informed decisions.
Despite previous efforts from the EU, large amounts of biodiversity data do not find their way into repositories due to a variety of reasons, including technical barriers. Many systems were not originally designed to handle the complexity of the biodiversity data being generated today. New methods and instruments, such as DNA-based observations, plankton imaging, passive acoustics, and biologging, can produce vast amounts of data and improve the collection and analysis of biodiversity time series. Nonetheless, the flow of these data into data repositories or integrators is at distinct stages of development and for some, the guidance is still a work in progress, subject to frequent changes as more information becomes available.
A data integrator refers to a facility that allows the storage of data and that provides tools that allow for the data to be analyzed. The terms integrator and aggregator are used interchangeably throughout the document. In contrast, a data repository is a facility that allows for data storage but does not provide functionalities for data analysis within the platform.

DTO-BioFlow is establishing data flows for several new biodiversity data types produced using different techniques and instruments which do not yet have established dataflows to make them available in long-term data repositories.
These include data from genomics observations, plankton imaging observations, fish, mammal and bird biologging, cetacean passive acoustic observations , and other relevant biodiversity data sources.
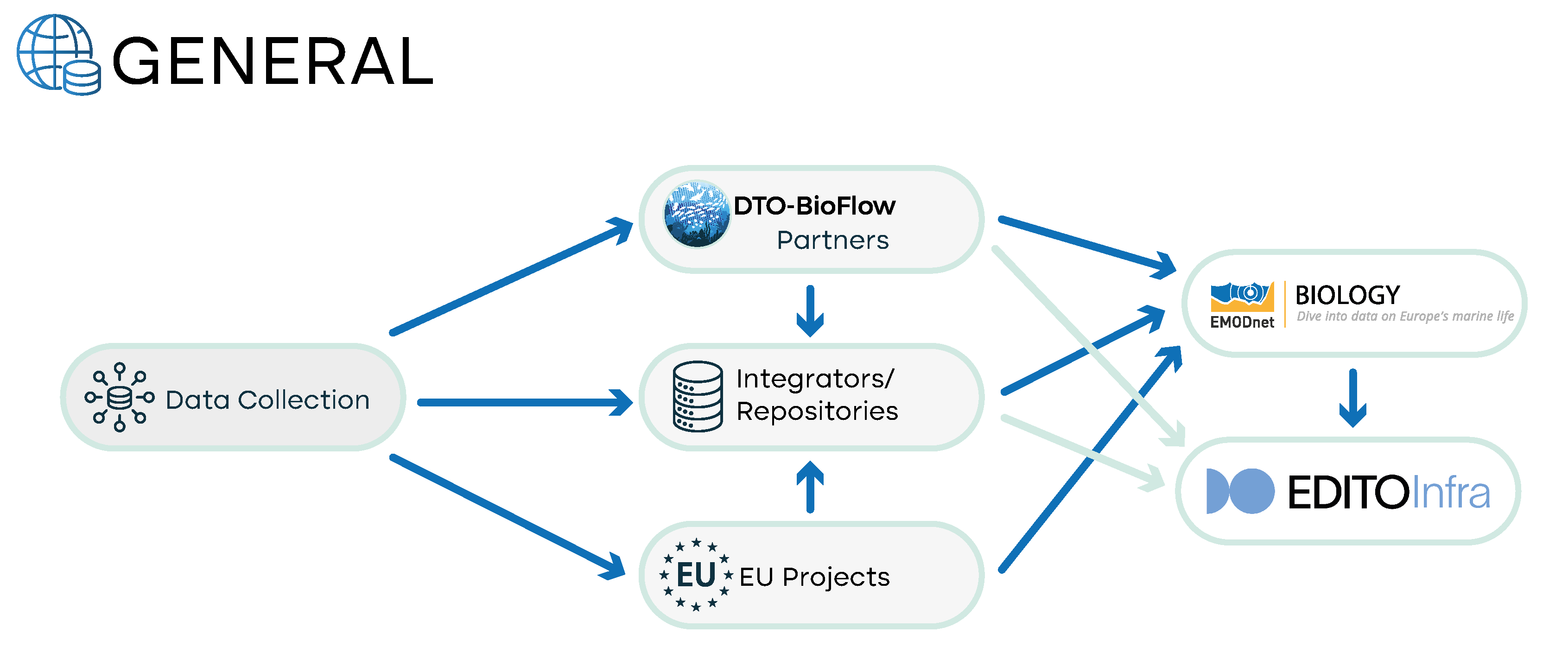
DTO-BioFlow contribution to EMODnet Biology
The European Marine Observation and Data Network (EMODnet) Biology is the European Union's service providing free and open access to in situ marine biodiversity data and data products. It is a data integrator that adheres to INSPIRE, Open Geospatial Consortium (OGC) standards for metadata and data and complies with the FAIR principles. EMODnet Biology uses the EurOBIS (European Ocean Biodiversity Information System) data infrastructure for providing data management. The two initiatives are therefore intrinsically connected and allow for data to reach a wider network of stakeholders using a common metadata sharing infrastructure with OBIS (Ocean Biodiversity Information System) and GBIF (Global Biodiversity Information System).
Within DTO-BioFlow, data pipelines for each of the data types addressed in the project will be established to maintain a sustained flow of biodiversity data towards EMODnet Biology, and ultimately into the EU DTO infrastructure.
The status of the data flows towards EMODnet Biology varies according to the diverse types of data addressed by DTO-BioFlow. In some cases, there is no data flow implemented, in other cases a data flow is already in place and will be subject to improvements during the project’s lifetime[1].


EU Digital Twin of the Ocean (EU DTO) infrastructure
The European Marine Observation and Data Network (EMODnet) Biology is the European Union's service providing free and open access to in situ marine biodiversity data and data products. It is a data integrator that adheres to INSPIRE, Open Geospatial Consortium (OGC) standards for metadata and data and complies with the FAIR principles. EMODnet Biology uses the EurOBIS (European Ocean Biodiversity Information System) data infrastructure for providing data management. The two initiatives are therefore intrinsically connected and allow for data to reach a wider network of stakeholders using a common metadata sharing infrastructure with OBIS (Ocean Biodiversity Information System) and GBIF (Global Biodiversity Information System). Within DTO-BioFlow, data pipelines for each of the data types addressed in the project will be established to maintain a sustained flow of biodiversity data towards EMODnet Biology, and ultimately into the EU DTO infrastructure. The status of the data flows towards EMODnet Biology varies according to the diverse types of data addressed by DTO-BioFlow. In some cases, there is no data flow implemented, in other cases a data flow is already in place and will be subject to improvements during the project’s lifetime[1].
Dowload the Blueprint Factsheet
What type of data will become available?
Genomic data is an umbrella term used to cover any kind of nucleotide sequence data, irrespective of whether it includes whole genomes, whole transcriptomes, or only specific loci. It therefore may refer to the nucleotide sequence information of individual organisms (e.g. for genetic barcoding), as well as of multiple organisms simultaneously by applying the prefix “meta-”. Whole genome sequence data of e.g. microbial community samples will consequently be called “metagenomic” data. In cases where multiple organisms are assessed simultaneously but the focus are individual loci (most commonly marker loci used for taxonomic identification of species), the term “metabarcoding” is used.
Genomic data may include taxonomic and/or functional annotations.
In the scope of this project, we mainly focus on taxonomically annotated nucleotide sequence data derived from environmental DNA (eDNA) samples subjected to either metabarcoding or metagenomic sequencing or both.
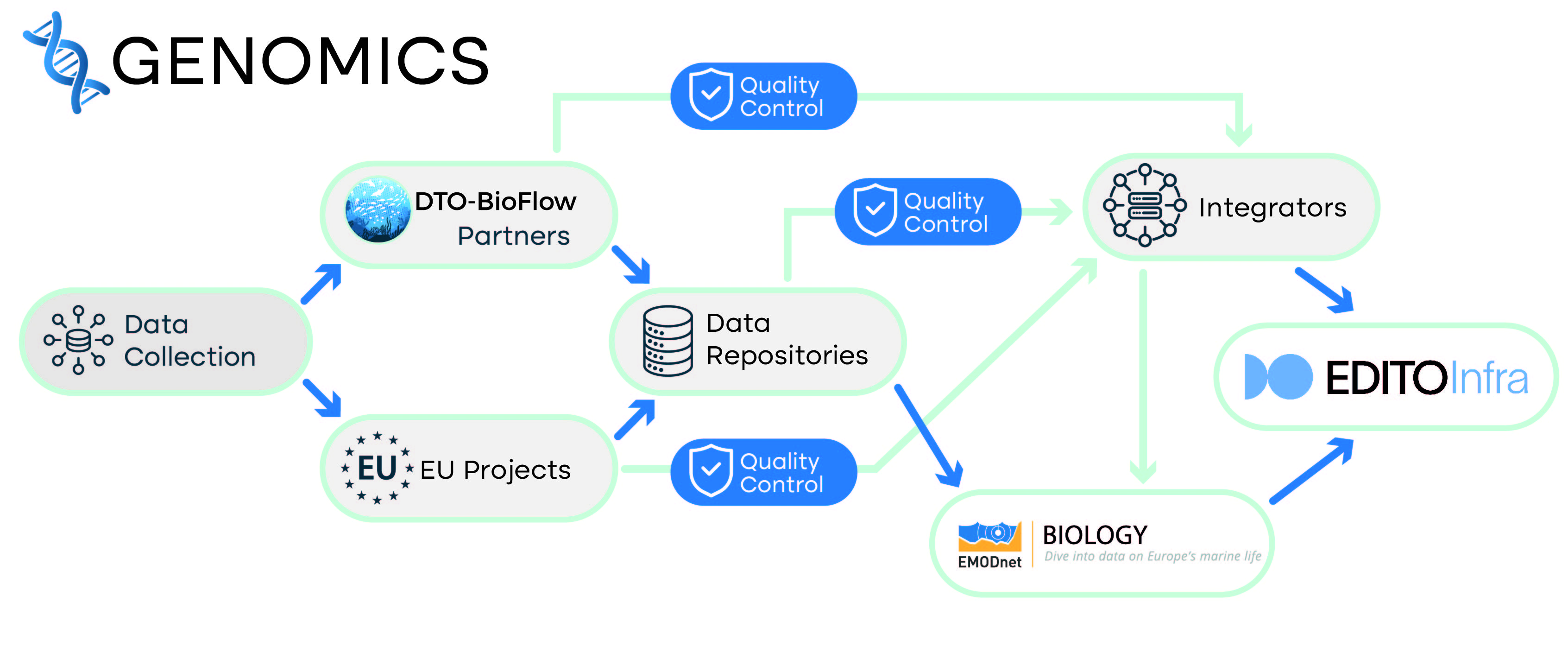
- Current status: there is no established genomics data flow to EMODnet Biology.
- EMODnet Data and Data Products Processing Level: L3 - Delayed mode data with further quality control, usually with some completeness, consistency and space/time uniformity. Data QC checks may include comparison with historical data and/or Level 5 products such as climatologies or gridded data.
- DTO-BioFlow upgrade: occurrence data of biological taxa in delimiter-separated tabular format, which will be annotated with literature information for species of interest, and environmental metadata, which can be retrieved from ENA BioSamples or from other repositories (e.g. PANGAEA).
- Related Data Project / Data repositories: MGnify (based on ENA), GBIF, OBIS
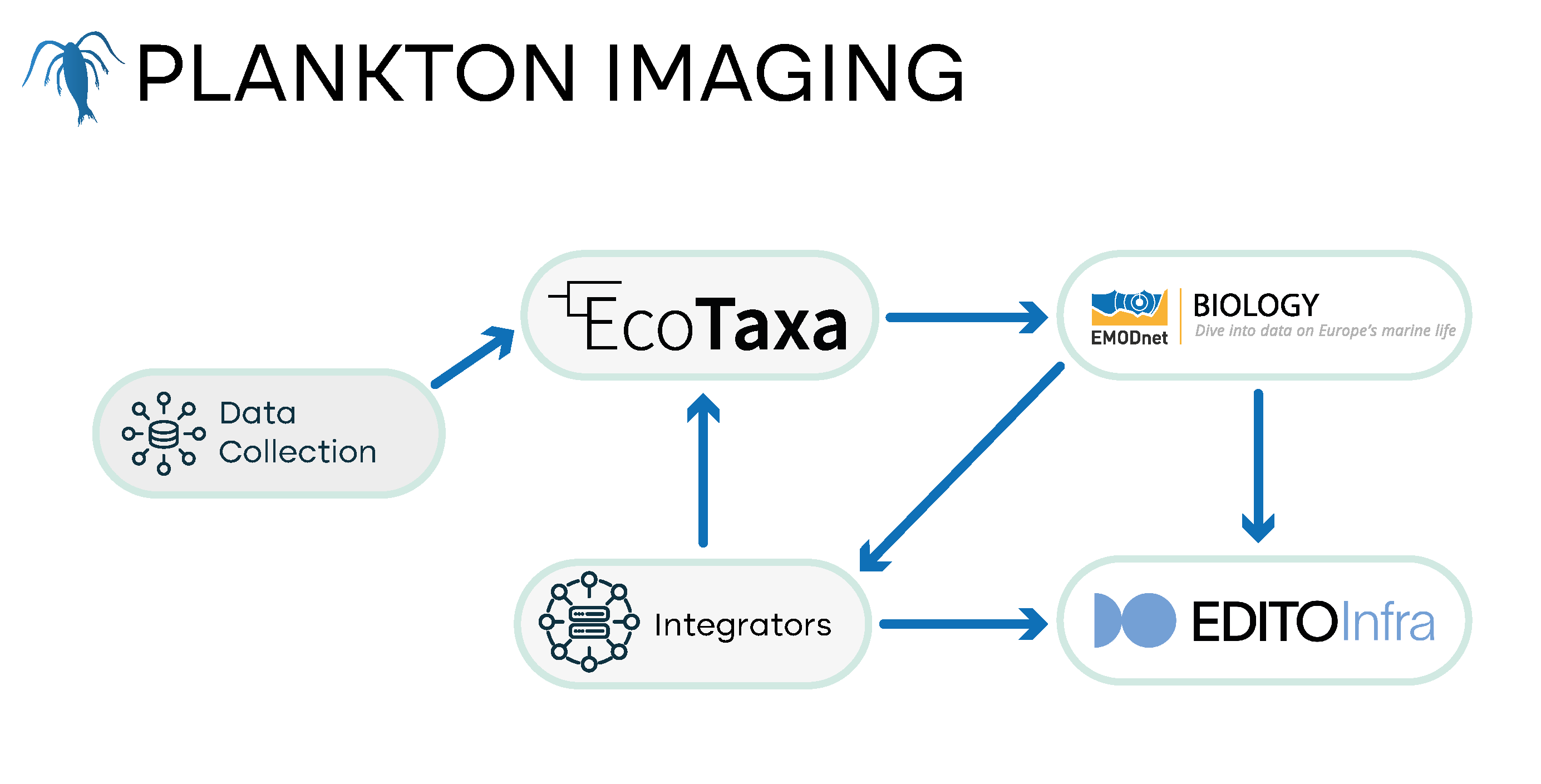
Plankton imaging data covers all data generated by quantitative imaging instruments - instruments that provide a large amount of images, in a consistent manner, to reliably extract quantitative information, such as concentrations and/or biovolumes. These observations of planktonic organisms allow us to better quantify their role as key trophic and functional links in open ocean ecosystems. In addition, the in situ instruments or samples are processed shortly after collection can provide information on so-called marine snow, which constitutes 80% to 90% of the particles in seawater. This marine snow plays a crucial role in marine ecosystems by facilitating the transfer of carbon to the deep ocean through the biological pump mechanism.
A wide variety of instruments can collect imaging data in situ or in the laboratory, each one with its own workflow. It is expected that data from plankton and particle imaging will flow into the EU DTO mostly via data integrators/repositories that are deployed across several in situ observatories and networks, notably including those overseen by DTO-BioFlow partners. The improved flows of data from plankton imaging expected from DTO-BioFlow will enhance biomonitoring efforts and global carbon flux estimations.
- Current status: After collection, images are processed, usually to extract Regions Of Interest (ROIs) from full frames and to measure them in numerous ways. While there is an existing flow of plankton imaging data to EMODnet Biology, several types of data from these images are not yet included in EMODnet Biology. This will be improved for various data integrators/repositories within the scope of the DTO-BioFlow project.
- EMODnet Data and Data Products Processing Level:
- L2 - Geo- and time-referenced processed (derived) data with a minimum QC. Near-real time (NRT) with full spatial and/or temporal resolution
- L3 - Delayed mode data with further QC, usually with some completeness, consistency and space/time uniformity. Data QC checks may include comparison with historical data and/or Level 5 products such as climatologies or gridded data.
- DTO-BioFlow upgrade: 3 data sets will be produced as part of WP4 Demonstrator Use Cases (DUC)
- microscopic count data from traditional monitoring; data from high-throughput imaging; next generation sequencing (DUC 4.3)
- biomass monitoring from stationary and on-demand mobile sensor platforms to assess and monitor biological processes like primary production or blooms and inform marine stakeholders (e.g., fisheries, DUC 6)
- maps of carbon export (EMODnet level 5 product) at global scale with some time resolution (e.g. seasonal) (DUC 7)
- Data repositories: EcoTaxa, EcoPart, PyOPIA, LifeWatch Observatory, SUBSIM
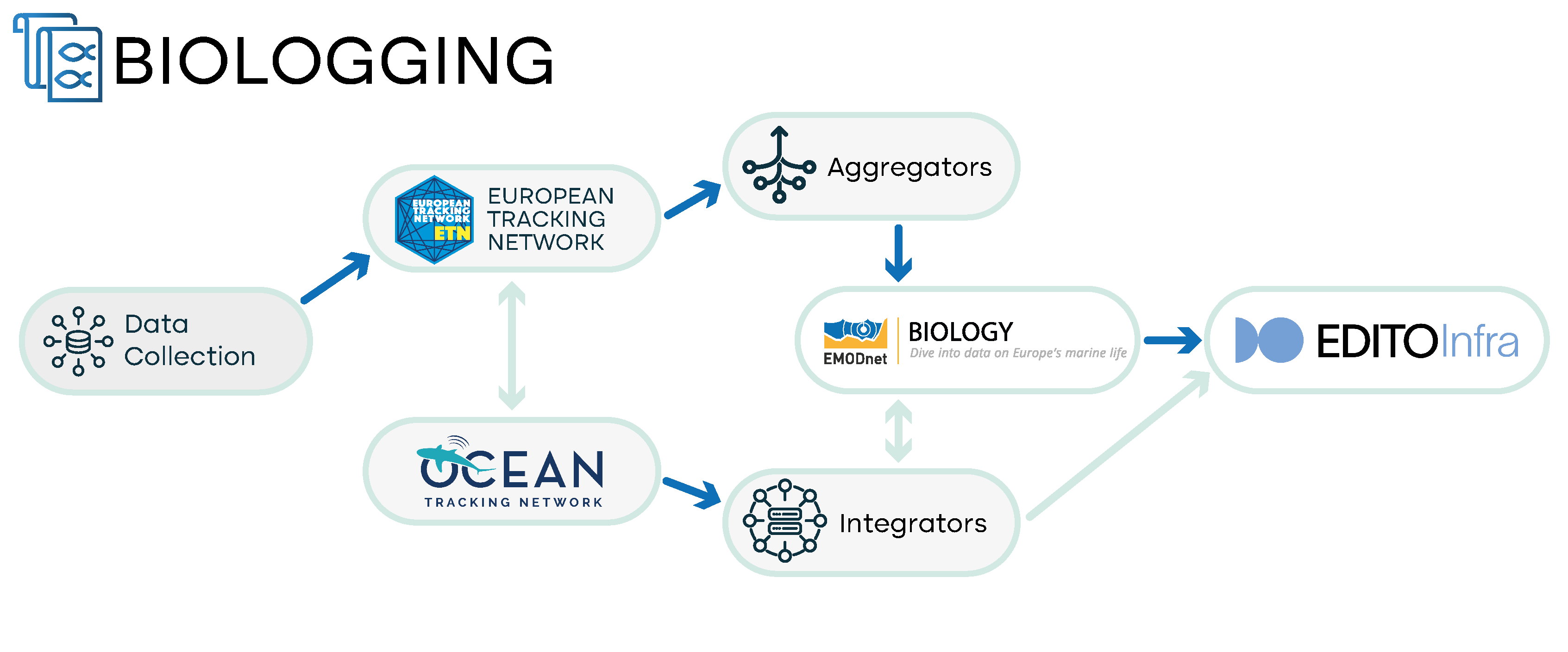
Biologging data are data of animal positions/presences obtained by animal-borne electronic devices. For this project, we consider biologging data from animals tagged in Europe, with mainly marine positions. They can be divided into three types:
- GPS tracking: these types of data involve electronic devices (such as GPS and SPOT tags) that record GPS positions. It is a technique that can be used for (marine) taxa with a large enough body size to carry a tag, and that live above or near the water surface so that a connection can be established with GPS satellites. Relevant taxa are large birds, marine mammals (cetaceans), fish (i.e. sharks) and marine reptiles (turtles). These types of data are managed in Movebank.
- Archival tracking: these types of data use electronic tags which store environmental data like water temperature, pressure (i.e. depth), acceleration, and light. The data is either downloaded from a detached and retrieved device (data storage tags) or directly sent through the ARGOS satellite system (e.g. pop-off satellite archival tags or PSATs). The trajectory of the animal can then be reconstructed through geolocation and hence requires processing of the raw data. These types of data can be managed in ETN.
- Acoustic telemetry: these types of data use a network of receivers that detect animal-borne acoustic transmitters when they pass in close vicinity of a receiver, a technique mainly used for fishes and crustaceans. The transmitters emit an acoustic signal (on a fixed frequency) with a unique ID on a predefined time interval and as the tagged animal moves through a network of receivers, it generates a trajectory. Detection range and therefore position resolution is 200m on average, albeit being highly dependent on prevailing environmental conditions. These types of data are managed in ETN.
- Current status: Biologging data are not operationally shared with EMODnet Biology yet, however data from biologging projects that are part of LifeWatch Belgium have been published as open data. These data were formatted as a Data Package and published as a Marine Data Archive for ETN, and a Zenodo deposit for Movebank. The latter GPS tracking data were also transformed and published as Darwin CoreArchive that are available in GBIF and OBIS and harvested from one of these aggregators by EMODnet Biology.
- EMODnet Data and Data Products Processing Level: None
- DTO-BioFlow upgrade: automated report files providing a summary of the tagged animals within a specific time frame or project, animal tracks and device maintenance reports. In addition, dynamic maps will be developed to map the detections or presences of tagged animals.
- Data repositories: Movebank, ETN
Passive Acoustic Monitoring (PAM) of sounds made by marine animals is an important method for estimating the distribution, density, and abundance of species that vocalise. It is particularly useful for animals that frequently produce species-specific vocalisations. Harbour porpoises (Phocoena phocoena) are excellent candidates as seldom does a minute go by without a porpoise producing species-specific echolocation clicks. The biological interpretation of acoustic data requires detecting signals produced by the animals, and the development and evaluation of detectors for classification is an active area of research. We are proposing the primary passive acoustic data to flow from ETN into EMODnet Biology.
- Current status: Currently, one dataset from passive acoustic monitoring can be found on EMODnet Biology for Phocoena phocoena from observation network in the Belgian Part of the North Sea. In addition, several PAM monitoring projects are collecting these data in the European seas which are currently not flowing to any data repository or aggregator. For the UK and North Sea waters a comprehensive inventory has been created, commissioned by the UK Department for Environment, Food and Rural Affairs. Most PAM data do not flow to EMODnet Biology or any other international data portal, except for the Baltic data that flows to the HELCOM repository. Metadata about the type of sensor and the settings used, and metadata about the deployment of the sensor are necessary but currently not included in the data available via EMODnet Biology. This metadata information should include the period of good quality recording, latitude, and longitude.
- EMODnet Data and Data Products Processing Level:
- L5 - Model or analysis output that uses data of Level 2 and/or 3 as input. Data products of this level represent the spatial distribution of a single parameter derived from multiple measurements. Data are aggregated and undergo some level of geo-processing and spatial or temporal interpolation to cover data gaps and/or solve data discrepancies.
- DTO-BioFlow upgrade: DTO-BioFlow will identify, compare and compile standards already used for observation, data exchange and reporting for specific species/species groups and specific habitats/regions by using existing data ingestion into HELCOM database as a starting point. We will also aim to develop script to generate maps
- Data repositories: HELCOM
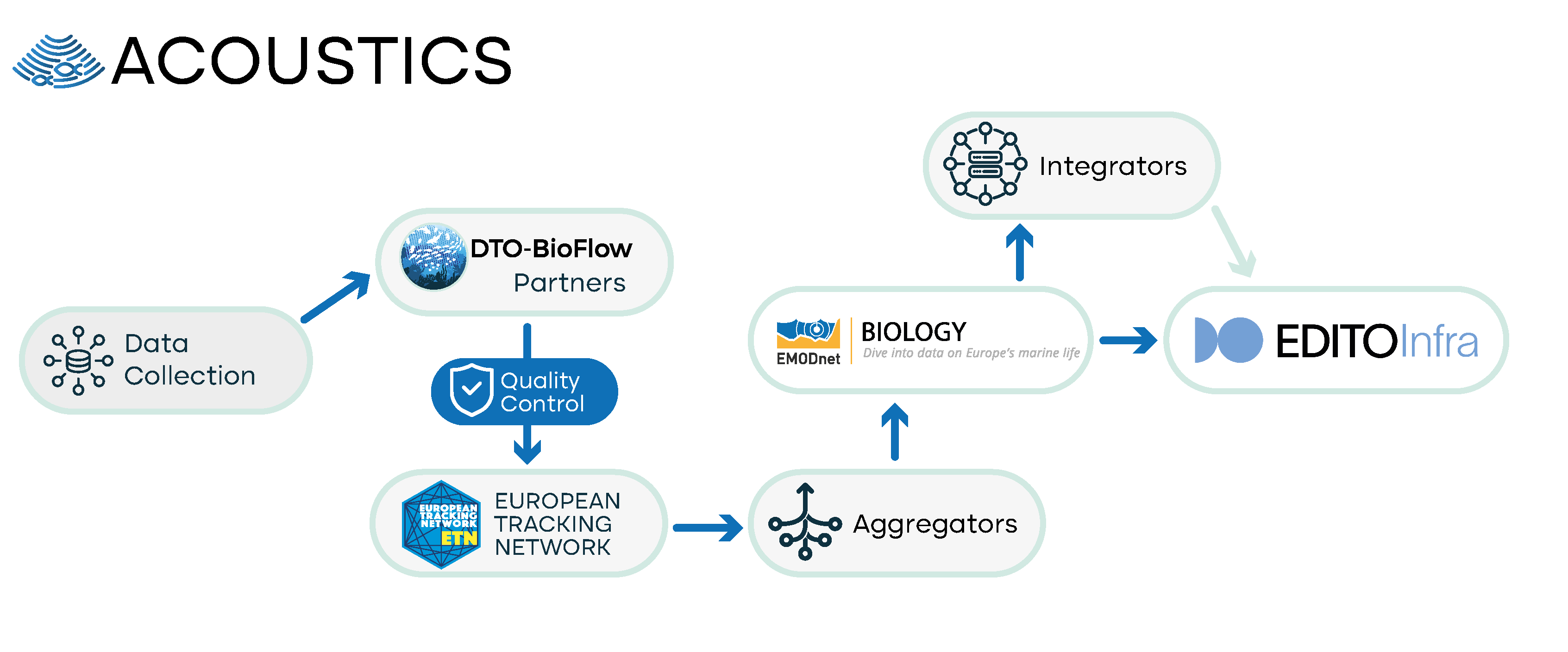
DTO BioFlow is considering other networks and data sources. These include species occurrence data from global platforms not yet integrated into EMODnet Biology, gridded species distributions from various projects, reporting data relevant to EU Directives, as well as data from industry, citizen science, and literature.
